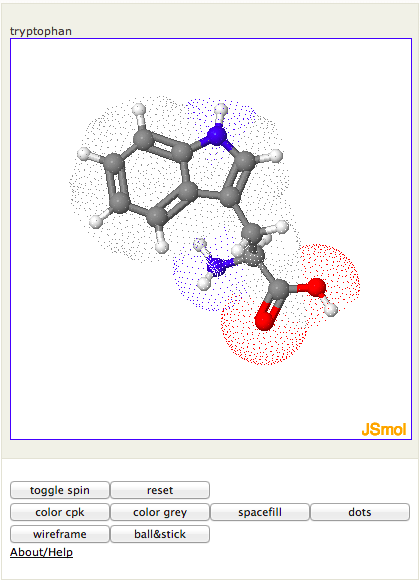
Figure 1. Screenshot of JSmol2wp viewer for a small molecule
This Wordpress plugin is based on the JSmol molecular graphics viewer from Bob Hanson, Zhou Renjian, and Takanori Nakane. Adaptation of JSmol to a Wordpress plugin was started by Jim Hu in June 2014. JSmol2wp should be considered beta!!! Use at your own risk and please report problems.
This is a shortcode extension. Use
[jsmol pdb='accession']
for a minimal version showing macromolecule structures from the PDB.
[jsmol pdb='accession']
is equivalent to
[jsmol acc='accession' type='pdb']

Figure 1. Screenshot of JSmol2wp viewer for a small molecule
Jmol can look up small molecules by name at the NCI resolver (http://cactus.nci.nih.gov/chemical/structure). Use
[jsmol acc = $name]where name is a molecule name. For example, acc='$tryptophan' loads the tryptophan structure (Figure 1).
Alternatively, you can use the NCBI PubChem server with:
[jsmol acc = :name]
If you use acc=accession and type=extension; JSmol2wp uses these to find the matching filename among the Wordpress uploaded media files. JSmol2wp will look to see if a file has been uploaded to your wordpress and it will use that file if it can find it. Example
[jsmol acc='caffeine' type='mol']
Will look for an uploaded file named caffeine.mol. If it can't find a matching post for an uploaded attachment and type=pdb, it will try http://rcsb.org/pdb. If it can't find a match there either, you'll get an error message in the JSmol window.
Alternatively, you can point JSmol2wp to a file path that is not in your media library. You can use
fileurl='http://path_to_file'
to force JSmol2wp to use that file. You can use any file with a publicly accessible URL
The captions parameter adds a text caption above the applet viewer.
JSmol2wp will always put a set of standard buttons below your viewer. It will try to guess which set of buttons to use, depending on whether you are loading a protein or a small molecule. This is determined by type (pdb or anything else)
The commands parameter allows you to run additional Jmol commands on loading or create buttons that will run commands. Separate commands with three pipes ( '|||")
Individual ommands have the format label=script, where label is the text you want on the button, and script is the Jmol script you want to run.JSmol2wp parses these, first by splitting on the ||| string and then on the = character. There are three kinds of command pairs.
add debug=value where the value can be:
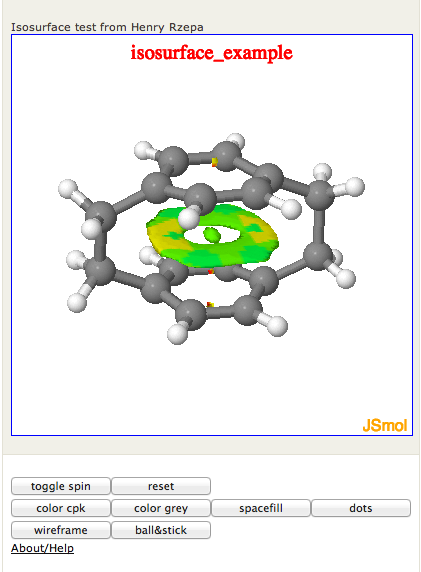
Figure 2. Screenshot of JSmol2wp viewer for a with an isosurface loaded from a local file
JSmol2wp can take additional optional parameters:
Figure 2 shows a structure and isosurface loaded from files uploaded to a local directory outside the Wordpress uploads path. The markup is:
[jsmol acc='isosurface_example' caption="Isosurface test from Henry Rzepa" fileurl='http://localhost/tmp/s_den.cub_.xyz' isosurface='http://localhost/tmp/s_den.cub_.jvxl']
Note that in this example, the acc parameter isn't really an accession. It's only being used to create a text label in the applet viewer.
[jsmol pdb='1LMB' caption='lambda repressor headpiece']
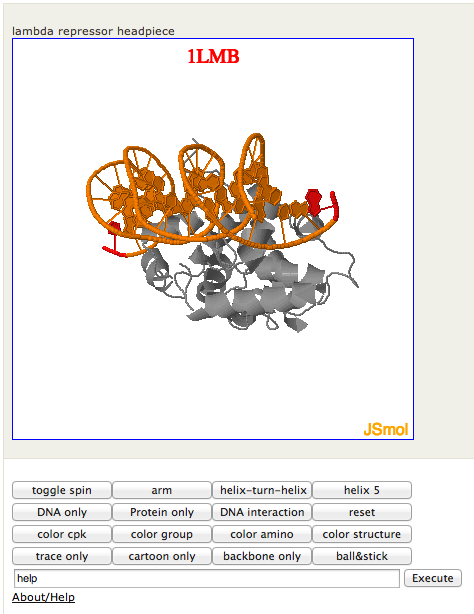
Figure 2. Screenshot of JSmol2wp viewer for a protein, with custom command/script execution buttons
Figure 3 is a screenshot of the sample viewer in this example post here. The markup for the lambda repressor example in that post is:
[jsmol pdb='1LMB' caption='λ repressor headpiece' commands=' arm = select 1-6:3, 1-6:4;color yellow;||| helix-turn-helix = select 33-52:3, 33-52:4; color yellow; hide hoh;||| helix 5 = select *:3,*:4; spacefill off; cartoon; select 84,87; color yellow; spacefill on; select *; hide hoh; hide *:1,*:2; #moveto 2 0 1 0 0 0 0; zoomto (84:4) 300;||| DNA only = display all; hide protein;||| Protein only = hide all; display protein;||| DNA interaction = hide all; display 33-52:3, 33-52:4, 1-6:3, 1-6:4, *:1, *:2; cartoon only; select 40-52:4; spacefill 23%; wireframe 0.15; color cpk; zoomto (45:4) 300; ']
This creates six custom buttons based on the text from the shortcode, and a reset button that tries to restore the original view. A lot of the suboptimal behavior of those buttons is from my lack of mastery of Jmol scripting. Note that Jmol commands can be used to load command sets from files.